This article and associated images are based on a poster originally authored by Nur M. Kocaturk, Andreas Holmqvist, Christina Duncan, Jennifer Riley, Steve Baginski, Graham Marsh, Joel Cresser-Brown, Hannah Maple, Kristiina Juvonen, Gajanan Sathe, Nicola Morrice, Calum Sutherland, Kevin Read and William Farnaby and presented at ELRIG Drug Discovery 2025 in affiliation with University of Dundee and Bio-Techne Ltd.
This poster is being hosted on this website in its raw form, without modifications. It has not undergone peer review but has been reviewed to meet AZoNetwork's editorial quality standards. The information contained is for informational purposes only and should not be considered validated by independent peer assessment.

Introduction
The conventional discovery of bifunctional targeted degrader molecules is generally reliant on the time consuming and expensive linear synthesis of 100s to 1000s of molecules. Furthermore, there is increased interest to identify degrader molecules capable of addressing central nervous system (CNS) therapeutic concepts. To rapidly access libraries of degrader molecules with diverse properties that may help us in the future identify the requirements for degrader brain penetration we present here a direct-to-biology (D2B) platform. This approach incorporates multiple types of conjugation chemistries within a 1-well-2-step protocol with sufficient conversion for direct cellular testing without the need of chromatography or protecting groups. We exemplify our approach using emerging E3 ligase and protein of interest (POI) ligands to produce bifunctional molecules with a diverse array of physicochemical properties, shapes, linker lengths and on-target degradation structure activity relationships. We here apply this approach to target Glycogen synthase kinase-3 (GSK3), a protein implicated in various CNS disease biology, including Alzheimer’s disease where it is known to regulate phosphorylation of Tau protein.
Direct-to-biology workflow
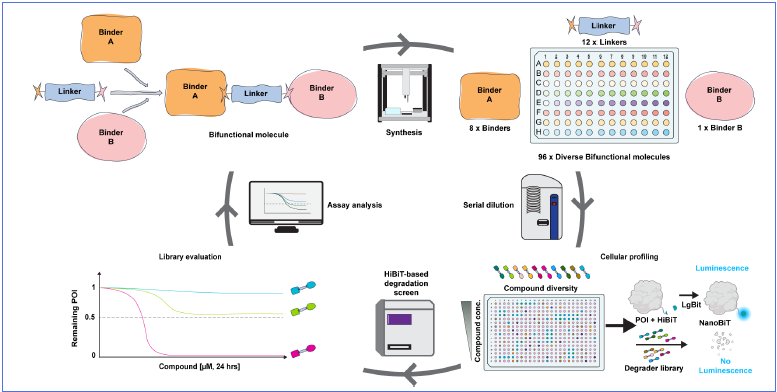
Image Credit: Image courtesy of Nur M. Kocaturk et al., in partnership with ELRIG (UK) Ltd.
Library synthesis approach
- Vary all components simultaneously e.g. E3 ligand, POI ligand and linker.
- Utilise orthogonally reactive linkers with high efficiency and reproducibility.
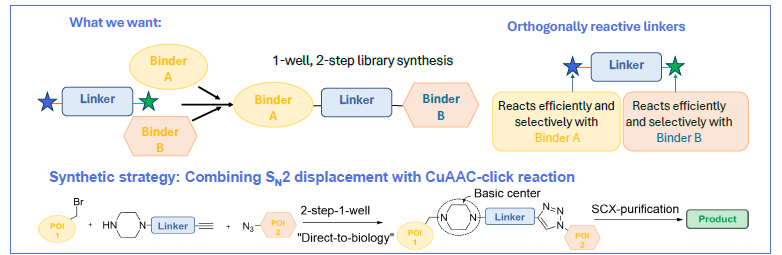
Image Credit: Image courtesy of Nur M. Kocaturk et al., in partnership with ELRIG (UK) Ltd.
Results
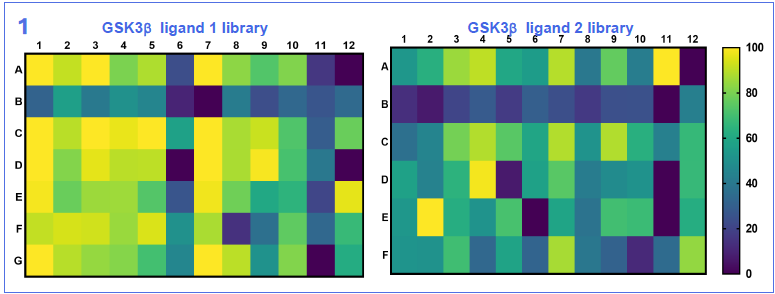
Image Credit: Image courtesy of Nur M. Kocaturk et al., in partnership with ELRIG (UK) Ltd.
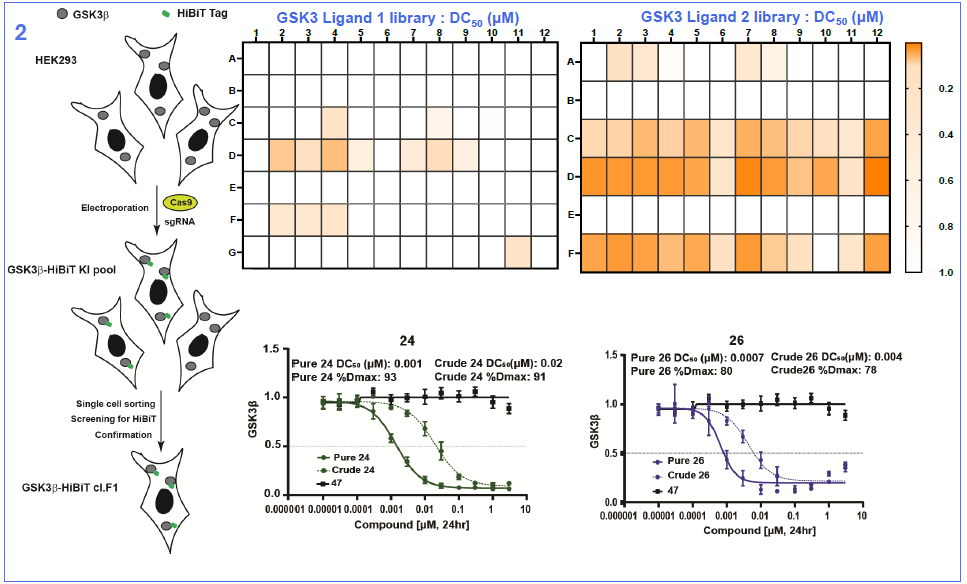
Image Credit: Image courtesy of Nur M. Kocaturk et al., in partnership with ELRIG (UK) Ltd.
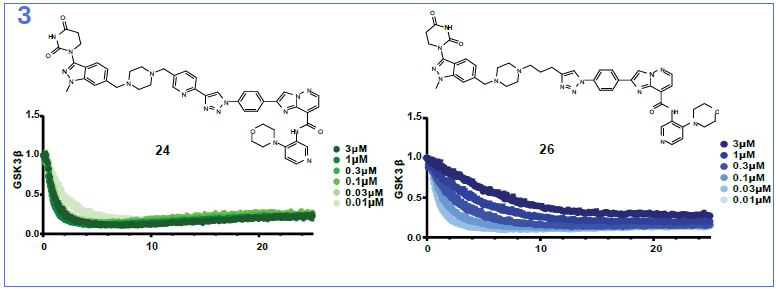
Image Credit: Image courtesy of Nur M. Kocaturk et al., in partnership with ELRIG (UK) Ltd.

Image Credit: Image courtesy of Nur M. Kocaturk et al., in partnership with ELRIG (UK) Ltd.
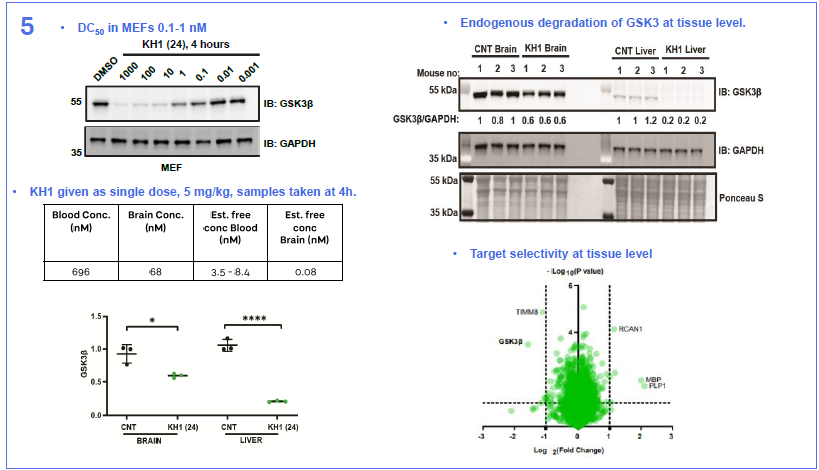
Image Credit: Image courtesy of Nur M. Kocaturk et al., in partnership with ELRIG (UK) Ltd.
Conclusion and future perspectives
In this study, we present a novel library synthesis and screening approach, that was able to identify an in vivo active bifunctional degrader of chemical probe quality directly coming from the initial screen. We started by establishing use of linker reagents that can be selectively conjugated in high conversion, with two different reaction types at either end, so-called orthogonally reactive linkers. Next, we took Direct-to-Biology approach to test our library in a GSK3β-HiBiT knock-in HEK293 cell line, generating full dose-response curves for all compounds. Once all building blocks and a suitable screening assay were available, we were able to complete the full synthesis and screening workflow in 5 days. We identified two molecules, KH1 (24) and KH2 (26) that showed DC50 values in the picomolar to single digit nanomolar range and exquisite specificity for degrading both GSK3 paralogs with no other significantly degraded proteins identified. Finally, we discovered that KH1 (24), following a single 5 mg/kg i.v. dose was able, within 4 hours, to almost completely remove GSK3β from the liver with partial degradation also observed in mouse brain. Our data support that this is achieved via a combination of moderate brain penetration and potent and rapid degradation kinetics. This study shows that the inherent pharmacokinetic/pharmacodynamic disconnect of PROTACs can be leveraged to avoid the requirement for high and prolonged CNS exposure, which would be required for inhibitors, to achieve on-target modulation in the brain. Whilst an opportunity for some, this may also act as an important warning to those who require avoiding on-target activity in the brain. We hope that the orthogonally reactive linker synthesis and screening approach exemplified here can provide a generalisable strategy to accelerate bifunctional probe discovery and therapeutic concept generation and a benchmark for CNS PROTAC discovery.
Acknowledgements
NMK is funded by EPSRC EP/X020088/1 and AH’s studentship is funded by EastBio BBSRC BB/T00875X/1 and BioTechne-Tocris.
About the University of Dundee
The University of Dundee is a public research university located in Dundee, Scotland, UK. Its core purpose is to transform lives locally and globally through the creation, sharing, and application of knowledge, underpinning its education, research, and community engagement activities.
About ELRIG (UK) Ltd.
The European Laboratory Research & Innovation Group (ELRIG) is a leading European not-for-profit organization that exists to provide outstanding scientific content to the life science community. The foundation of the organization is based on the use and application of automation, robotics and instrumentation in life science laboratories, but over time, we have evolved to respond to the needs of biopharma by developing scientific programmes that focus on cutting-edge research areas that have the potential to revolutionize drug discovery.
Comprised of a global community of over 12,000 life science professionals, participating in our events, whether it be at one of our scientific conferences or one of our networking meetings, will enable any of our community to exchange information, within disciplines and across academic and biopharmaceutical organizations, on an open access basis, as all our events are free to attend!
Our values
Our values are to always ensure the highest quality of content, that content will be made readily accessible to all, and that we will always be an inclusive organization, serving a diverse scientific network. In addition, ELRIG will always be a volunteer-led organization, run by and for the life sciences community, on a not-for-profit basis.
Our purpose
ELRIG is a company whose purpose is to bring the life science and drug discovery communities together to learn, share, connect, innovate and collaborate, on an open access basis. We achieve this through the provision of world-class conferences, networking events, webinars, and digital content.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.
Last Updated: Jan 6, 2026